High Q/V factors and light squeezing in a wavelength scale volume has been a challenging goal for a long time. In this context, we worked out the integration of photonic bandgap nanocavities in sub-micron sized waveguides. By the means of new geometries reducing modal mismatch losses at cavity-mirrors interfaces, the lab contributed to the advent of so called nanobeam cavities and the demonstration of record Q factors. In the previous period, we established the operation of on-chip SOI optical tweezers and achieved single nanoparticle trapping as well as multiple particles assembly through near field interaction with nanobeam cavities. By wavelength induced field map tuning within a set of coupled nanobeam cavities, we also evidenced particle motion control and handling between specific trapping sites. Since then, we have moved one step further introducing opt fluidic taming of a colloidal dimer
[1] as well as lateral motion along few-mode silicon waveguides
[2]. We also evidenced the near-field mapping of the electromagnetic field around a nanobeam cavity with trapped microbeads
[3].
We then decided to move towards the “bio-world”, a completely new area for the lab. Thus we started to tackle the challenge of biological species. Even if micron-scale objects might be commonly handled in free space through a microscope working as an optical tweezer, we suggested that the lower powers involved in nanoscale tweezers would particularly suit to the manipulation of biological objects as bacteria. Furthermore, since the field of bacterial antibiotic resistance emerged as a major societal challenge, we decided to develop an approach based on optical trapping of bacteria with SOI nanobeam cavities.
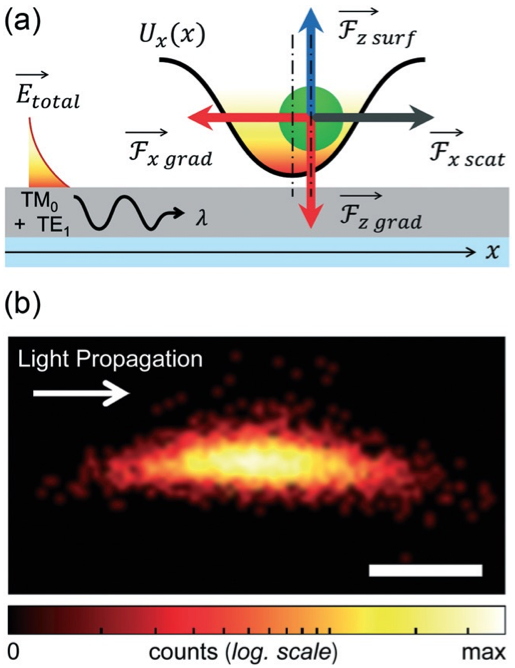

Figure 1: Left (a) Schematic representation of the forces acting on a particle trapped in a TM0–TE1lattice. (b) Experimental map of the trapping potential experienced by a 1μm bead in a TM0–TE1 lattice (scale bar: 250nm).
Right: (a) principle of the optical trapping experiment: the photonic crystal structure is immersed in water (1) thanks to a PDMS frame that allows for the Brownian motion of bacteria in the proximity of the optical cavity. Light from a tunable laser is injected at the resonance wavelength into the waveguide in an end-fire setup. The transmitted light is collected from the other facet of the waveguide via a microscope objective and its intensity is monitored with an oscilloscope. (b) SEM picture of a bacterium used for the experiment (c) SEM picture of the nanobeam cavity located at the middle of the 10mm long monomode waveguide.
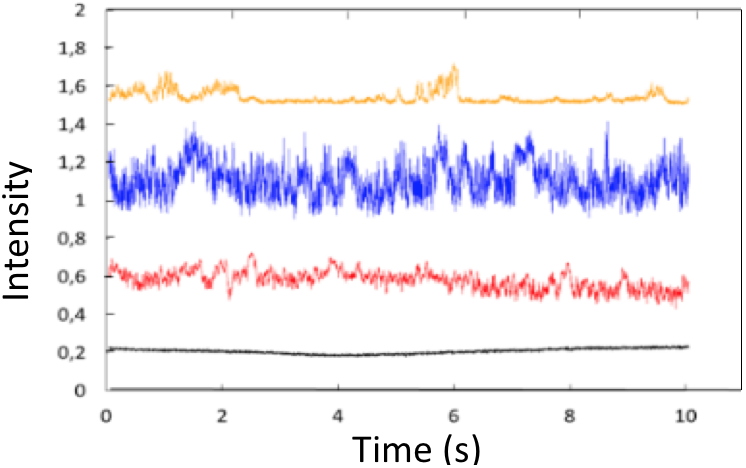
We made the very first demonstration of the optical trapping of a bacterium by a SOI nanobeam cavity
[4]. Then the spatial analysis of the Brownian motion of a bacterium in the trap, correlated with the signal transmitted by the optical structure allowed an accurate characterization of the behavior of the trapped object. The trapping signal of these bacteria transmitted through the optical structure reveals indeed two mechanisms. First, as long as the bacterium is trapped by the cavity resonant field, the bacteria cavity optical interaction leads to a shift of the cavity resonant wavelength. At a fixed observation wavelength it translates into a sudden variation of the transmitted intensity. Second, strong oscillations of the transmitted intensity at the very same wavelength are observed, again while trapping of the bacterium. These oscillations appeared not to be noise but indeed related to small displacements of the bacterium within the evanescent field above thenanobeam cavity. Furthermore, the analysis of the fluctuations of the transmitted signal as a function of time during trapping made it possible to demonstrate a proper signature for different types of trapped bacterium. Using this method, we showed that the combination of these two characteristics allows the absolute identification of the studied bacteria. We thus demonstrated the trapping and identification of three bacteria:
S. epidermidis,
E. coli and
B. subtilis. through the spatial and temporal characterization of their trapping above a nanobeam cavity
[4].

Figure 2: (1) Trapping Setup and SEM images of the under study bacteria: (a) diplococcus of
S. epidermidis, (b)
E. coli, and (c) aggregate of
B. subtilis; (2) Global shape of trapped bacteria trajectories on the SOI microcavity; (3) Example of intensity transmitted through the microcavity during 10s of trapping for the bacteria
S. epidermidis diplococcus (orange),
E. coli (blue),
B. subtilis aggregate (red) and with no bacteria trapped (in bacteria and water medium) (black).
We then further extend this approach to the demonstration of the optical trapping of bacteria in a 2D silicon hollow photonic crystal cavity and discovered that this geometry allowed us for the Gram-type differentiation of bacteria at the single cell scale, in a fast, label-free, and non-destructive way. A bacterium is Gram-positive or Gram-negative following its reaction to a staining test called the Gram test. This is the first test to be performed when trying to identify the bacterium responsible for an infection during a bacterial diagnosis. The Gram type translates also into different membrane structures. In collaboration with the EPFL (Switzerland), the trapping of seven types of living bacteria has been demonstrated, species featuring different morphologies, motilities (presence or absence or flagella), and Gram staining properties:
Pseudomonas putida,
Neisseria sicca,
Escherichia coli,
Yersinia ruckeri,
Bacillus subtilis,
Listeria innocua and
Staphylococcus epidermidis
[5].
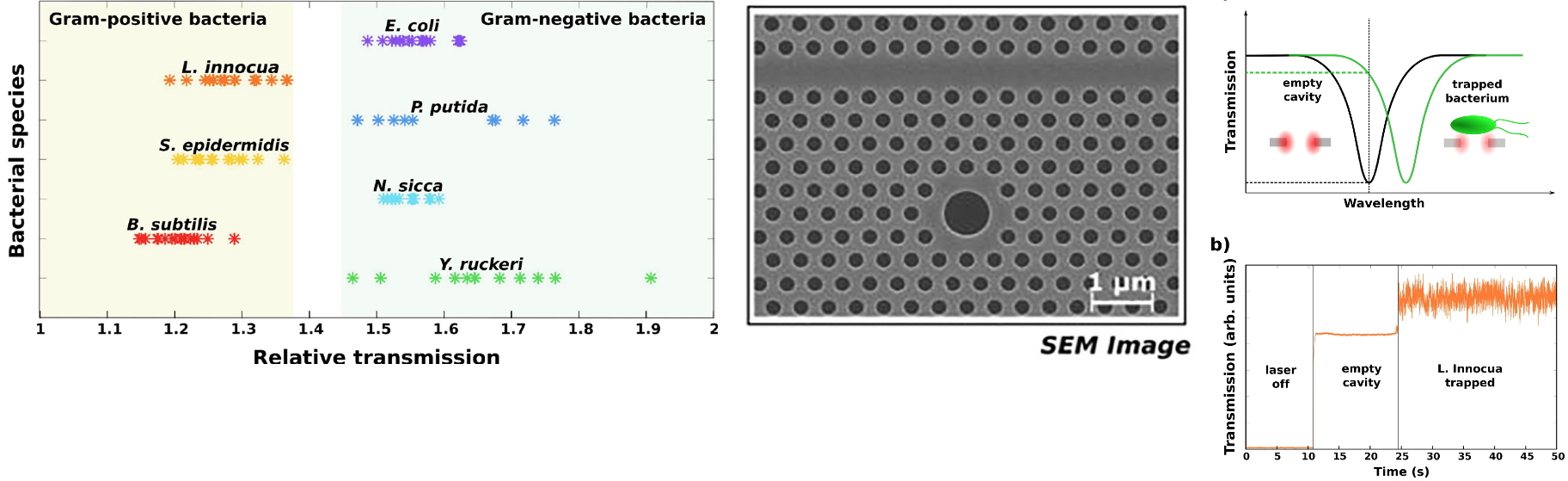
Figure 3: Left - SEM image of the 2D hollow photonic crystal SOI cavity.
Middle (a): cavity shift induced by the trapped bacterium, (b): transmission offsets induced by the trapped bacterium,
Right: relative transmission for different bacteria species allowing gram type identification.
As illustrated above, this approach makes it possible to identify the Gram type of bacteria by a fast label-free method, working at a low concentration of bacteria. Moreover, this method is non-destructive, unlike the classical Gram test, it is therefore possible to use the same cells to perform further analyses, promoting their identification.
Today bacteria are an important subject of study in the biomedical sector because they are a cause of many infections in the human body. The current diagnostic procedures are mainly based on the culture of bacteria populations in Petri dish which allow us to identify the bacterium responsible for the infection and the antibiotics that will treat it in 48 hours. In order to reduce this time of diagnosis and lower the use of wide spectrum generic antibiotics, we attempted to work on bacteria identification at the single cell level by exploiting optical forces and exploring cavity bacteria optical interactions.
 References
[1] Pin C, Cluzel B, Renaut C, Peyrade D, Picard E, Hadji E and De Fornel F
References
[1] Pin C, Cluzel B, Renaut C, Peyrade D, Picard E, Hadji E and De Fornel F Optofluidic taming of a colloidal dimer with a silicon nanocavity.
Applied Physics Letters, 2014,
105(17): 171108
[2] Pin C, Jager JB, Tardif M, Picard E, Hadji E, De Fornel F and Cluzel B
Optical tweezing using tunable optical lattices along a few-mode silicon waveguide.
Lab on a Chip, 2018,
18: 1750-1757
[3] Pin C, Cluzel B, Renaut C, Picard E, Peyrade D, Hadji E and Fornel
Optofluidic near-field optical microscopy: Near-field mapping of a silicon nanocavity using trapped microbeads.
ACS Photonics, 2015,
2(10): 1410-1415
[4] Tardif M, Jager JB, Marcoux P, Uchiyamada K, Picard E, Hadji E and Peyrade D
Single-cell bacterium identification with a SOI optical microcavity.
Applied Physics Letters, 2016,
109: 133510
[5] Therisod R, Tardif M, Marcoux PR, Picard E, Jager JB, Hadji E, Peyrade D and Houdré R
Gram-type differentiation of bacteria with 2D hollow photonic crystal cavities.
Applied Physics Letters, 2018,
113: 111101