Bacteria are an important subject of study in the biomedical sector because they are a cause of many infections in the human body. The current diagnostic procedures are mainly based on the culture of bacteria populations in Petri dish which allow us to identify the bacterium responsible for the infection and the antibiotics that will treat it in 48 hours. In order to reduce this time of diagnosis we propose to work on the analysis of bacteria at the level of the single cell by exploiting optical forces.
They are generally used in open space through a microscope, as optical tweezers. However, other optofluidic devices have recently emerged, based on the use of integrated on-chip optical components such as photonic crystals and microcavities. The low powers involved in these new devices are particularly suited to the manipulation of biological objects.
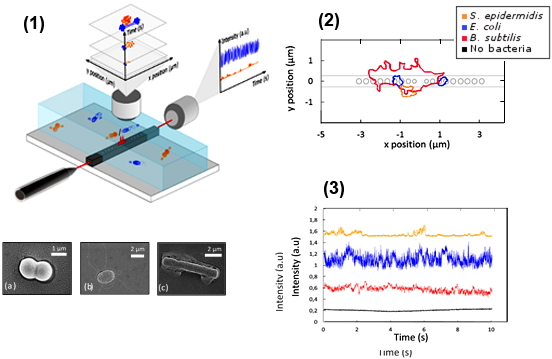
Figure : (1) Trapping Setup and SEM images of the under study bacteria: (a) diplococcus of
S. epidermidis, (b) E. coli, and (c) aggregate of
B. subtilis; (2) Global shape of trapped bacteria trajectories on the SOI microcavity; (3) Example of intensity transmitted through the microcavity during 10s of trapping for the bacteria
S. epidermidis diplococcus (orange),
E. coli (blue),
B. subtilis aggregate (red) and with no bacteria trapped (in bacteria and water medium) (black).
We developed an optical trapping device based on the use of SOI microcavities and dedicated to the study of bacteria. The spatial analysis of the Brownian motion of a bacterium in the trap, correlated with the signal transmitted by the optical structure allows an accurate characterization of the behavior of the trapped object. Moreover, the analysis of the fluctuations of the transmitted signal as a function of time during the trapping makes it possible to demonstrate the signature of each type of trapped bacterium. Using this method, we have demonstrated the trapping and identification of three bacteria:
S. epidermidis,
E. coli and
B. subtilis. We will now extend this study of bacterial / electromagnetic field interactions to the discrimination of different bacterial genera or species. We also intend to correlate the optical signature of a trapped bacterium to external events (life and death, cell division).
The optical structures used in this project are fabricated in the PTA by J-B. Jager.